Cancer Biomarker Discovery

Therapeutic biomarker discoveries rely on “big” and “reliable” data resources. Identifying disease-specific target genes and their particular associations with clinical factors remain challenging. This challenge arises not only owing to the property of high dimensions vs. low sample size in genomic data, but also through long-standing confusion in the literature occasioned by inconsistent discoveries of differing types of data resources across studies. By re-organizing and re-analyzing existing cancer genomics and clinical data, we have identified novel, clinically relevant gene targets and experimentally have validated their functions. These identified targeted genes or gene signatures are potential therapeutic biomarkers for personalized diagnosis and treatment.
pathway and gene network analysis
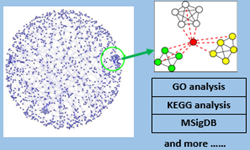
Cancer development involves a group of gene interactions arising through complex regulatory mechanisms. A common way to understand these underlying mechanisms is by integrative analysis, which considers the whole organism as a system. Integration of heterogeneous cancer biological data by interdisciplinary collaborations is critically important to address cancer questions through the combination of experimental and computational analysis from a systems biology perspective. The success of pathway and biological function analysis based on gene network database mining. By developing computational methodology and in vitro experimental validation, we were able to identify interestingly epigenetic targets and explain their functional mechanism from a systems biology perspective.
Drug response predictions

The identification of molecularly targeted drugs that response to the cancer treatment is the most exciting field of cancer therapeutics, as those drugs provide dramatic clinical benefits with little toxicity. The development of reliable machine learning algorithms (e.g. regression, classification and clustering models) is of critical importance in the accuracy of drug response prediction. Based on information now being collected from the laboratory and public database, we developed novel statistical models to identify the most “sensitive” and "responsive" anticancer drugs that have been proved to be effective in eradicating human tumor cells, which will benefit to personalized medicine.
Genetic evolutionary study on microbiomes
We are also interested in address evolutionary and genetic association problems among microbiomes. Some microbes, such as plasmid and bacterial microbes, have genomes that accept DNA from foreign species or their environment. However, owing to random gene recombination and mutation, along with adaptation to environmental changes, it is very challenging to identify microbe linkages based on their DNA. I developed new methods for relating a large number of bacterial or plasmids based on their genetic sequences by using a computational approach analogous to the hybridization of "mixed-genome” microarrays. Our publications associated with this research showed that these novel classification methods successfully deal with microbes’ genetic association and linkage relationship problems.